RecPhyloXML
XML for reconciled gene trees
Introduction
recPhyloXML and recGeneTreeXML are two XML grammars inherited from phyloXML [1] designed to describe reconciled gene trees. They were created in order to allow interoperability between the various scripts and softwares used in reconciled gene trees studies.
Fundamental implementation
recGeneTreeXML grammar allows you to add a new tag <eventsRec> in phyloXML <clade>. This tag describes the different evolutionary events associated to this clade. To distinguish phyloXML trees
from reconciled gene trees inferred by a reconciliation process, the root tag <phyloxml> is replaced by <recGeneTree>.
recPhyloXML grammar allows you to store and share one or more reconciled genes trees with the associated species tree.
- > Reconciled gene trees have to be described in recGeneTreeXML grammar.
- > The species tree has to be described in phyloXML grammar.
recGeneTreeXML
Presentation
recGeneTreeXML enriches phyloXML vocabulary by adding the complex tag <eventsRec>. This tag is situated in the authorized tag sequence of the <clade> tag (voir documentation).
The content of the <eventsRec> tag represent the sequence of evolutive events happening along the branch of a gene tree, including the position of these events on the associated species tree. These events are inferred
during a complex process called reconciliation[2] .
The gene history events that can be included in the <eventsRec> can be classified in two groups:
-
> Non terminal events:
<transferBack>. This tag can be used as many times as necessary and in any order. This event does not cause any bifurcation in the gene tree and is supposed to happen along one of its branch. -
> Terminal events:
<speciation>,<branchingOut>,<bifurcationOut>,<duplication>,<loss>and<leaf>. There is exactly one of these tag at the end of the sequence contained in a<eventsRec>tag.
The terminal events describe events causing either a bifurcation in the gene tree (<speciation>,<branchingOut>,<bifurcationOut>,<duplication>) or the end
of a lineage (<loss> and
<leaf>). In any case, they mark the event associated with the extremity of a gene tree branch.
Events description
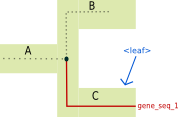
<leaf> tag:
The <leaf> tag indicates that the branch ends on a gene tree leaf.
Illustrated example
Associated recGeneTreeXML code
<clade>
<name>gene_seq_1</name>
<eventsRec>
<leaf speciesLocation="C"></leaf>
</eventsRec>
</clade>
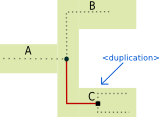
<duplication> tag:
The <duplication> tag represents a gene duplication inside a species.
Illustrated example
Associated recGeneTreeXML code
<clade>
<eventsRec>
<duplication speciesLocation="C"></duplication>
</eventsRec>
</clade>
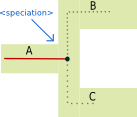
<speciation> tag:
The <speciation> tag describes a gene indergoing a bifurcation because the species in which it evolves speciates.
Illustrated example
Associated recGeneTreeXML code
<clade>
<eventsRec>
<speciation speciesLocation="A"></speciation>
</eventsRec>
</clade>
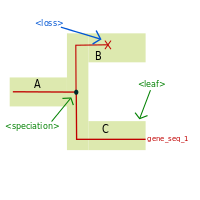
<loss> tag:
the <loss> tag describes the loss of a gene lineage. As a lost gene has no children, a <clade> containing a <loss>
tag should not contain any <clade> children.
Usually, this tag will appear in one of the children of a clade that underwent a speciation.
Illustrated example
Associated recGeneTreeXML code
<!--Example with end tag <leaf> -->
<clade>
<name>gene_seq_1_ancestor</name>
<eventsRec>
<speciation speciesLocation="A"></speciation>
</eventsRec>
<clade>
<name>gene_seq_1</name>
<eventsRec>
<leaf speciesLocation="C"></leaf>
</eventsRec>
</clade>
<clade>
<name>gene_seq_1_lost</name>
<eventsRec>
<loss speciesLocation="B"></loss>
</eventsRec>
</clade>
</clade>
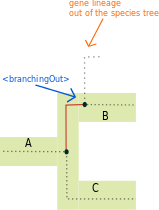
<branchingOut> tag:
The <branchingOut> tag represents a event where a gene lineage splits and one copy exits the species tree branch while the other gene copy remains in the species branch. They are typically part of an horizontal gene transfer event.
Illustrated example
Associated recGeneTreeXML code
<clade>
<eventsRec>
<branchingOut speciesLocation="B"></branchingOut>
</eventsRec>
</clade>
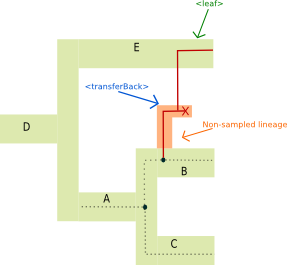
<transferBack> tag:
The <transferBack> tag represents an horizontal gene transfer toward a branch of the species tree.
Illustrated example
Associated recGeneTreeXML code
<!--Example with end tag <leaf> -->
<clade>
<eventsRec>
<transferBack destinationSpecies="E"></transferBack>
<leaf speciesLocation="E"></leaf>
</eventsRec>
</clade>
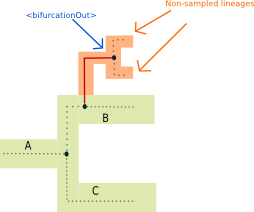
<bifurcationOut> tag:
The <bifurcationOut> tag represents a bifurcation in the species tree that would happen while the gene evolves in a species that is not represented in the species tree (following a model implicating non-sampled / extinct lineages that are absent from the species tree [3]).
Illustrated example
Associated recGeneTreeXML code
<clade>
<eventsRec>
<bifurcationOut></bifurcationOut>
</eventsRec>
</clade>Lateral Gene Transfer
A lateral gene transfer is represented in two steps, with their respective tags: <branchingOut> and <transferBack>.
The <branchingOut> tag represent the action of leaving a species branch, it specifies where the transfer comes from (through the speciesLocation attribute).
The <transferBack> tag represent the action of entering a species branch, it specifies where the transfer arrives in (though the destinationSpecies).
Event tags attributes
Aside from the <bifurcationOut> and <transferBack> tag, all event tag have an obligatory speciesLocation attributes, which specifies in which species the event takes place. For <bifurcationOut>,
the event always take place in a non-sampled / extinct lineage. <transferBack> events have instead a destinationSpecies attributes which contains the species that receive the transfer. Additionally, all event
tag have a facultative timeSlice attribute that can, in models where the species tree is subdivided for instance, provide information on the timing of the event. Finally, the <leaf> tag has a facultative geneName
attributes that can specify which extant gene it corresponds to.
recPhyloXML
Presentation
recPhyloXML facilitates the exchage of several gene family that were reconciled to a same gene tree. Its structure is fairly simple.
A <recPhylo> root tag contains the following sequence:
- > 0 .. 1 species tree. phyloXML format . BUT contains in the
<spTree>tag rather than the<phyloxml>tag and whose clade can possess the<geography>tag (see below) - > 1 .. n gene family tree. recGeneTreeXML format. Each contained in a
<recGeneTree>tag
annotation with geographical information
Geographical annotations can be indicated for reconciled gene and species tree <clade> tags thanks to the <geography> tag. The geographical annotation mainly consists in an area, KML information for displaying areas in GIS software and geographic information as defined in the usual PhyloXML grammar.
A <geography> tag contains the following sequence:
- > 1
<area>tag, described below - > 0 .. 1 location described using the Placemark standard of OpenGeoSpatial KML . Contained in a
<KML_location>tag - > 0 .. 1 location described using the Distribution standard of phyloXML. Contained in a
<phyloXML_location>tag
An area (<area>) contains the following elements:
- > 1
<name>, which names the area - > 0 .. 1
<desc>, a description - > 0 .. 1
<value>, such as a support (must be a number) - > 0 .. 1
<source>(e.g. "observed" or "inferred by Beast")
<geography> tag :
<clade>
<name>gene_seq_1</name>
<eventsRec>
<leaf speciesLocation="C"></leaf>
</eventsRec>
<geography>
<area>
<name>locationFR67160</name>
<desc>a lovely town</desc>
<value>87</value>
<source>input data</source>
</area>
<KML_location>
<name>locationFR67160</name>
<Point>
<coordinates>49.0320419,7.9534841,158</coordinates>
</Point>
</KML_location>
<phyloXML_location>
<desc>a lovely town</desc>
<Point geodetic_datum="WGS84">
<lat>49.0320419</lat>
<long>7.9534841</long>
<alt>158</alt>
</Point>
</phyloXML_location>
</geography>
</clade>
Files
- .xsd files: get them here
- recGeneTree Example: Download here
- recPhyloXML Example: Download here